Bacteria are everywhere — in soil, water, air, on our skin, and inside our bodies. While many are harmless or even beneficial, others can cause devastating diseases, spoil food, or disrupt ecosystems. Identifying bacteria accurately is not just a laboratory curiosity; it's a cornerstone of modern medicine, public health, agriculture, and biotechnology. From diagnosing infections to ensuring safe drinking water, the ability to pinpoint bacterial species transforms uncertainty into actionable knowledge.
The science of bacterial identification has evolved dramatically over the past century. What once relied solely on staining techniques and growth patterns now integrates genomics, artificial intelligence, and rapid diagnostics. Understanding why this process matters — and how it’s done — empowers healthcare providers, scientists, and policymakers to make informed decisions that save lives and protect environments.
The Critical Importance of Bacterial Identification
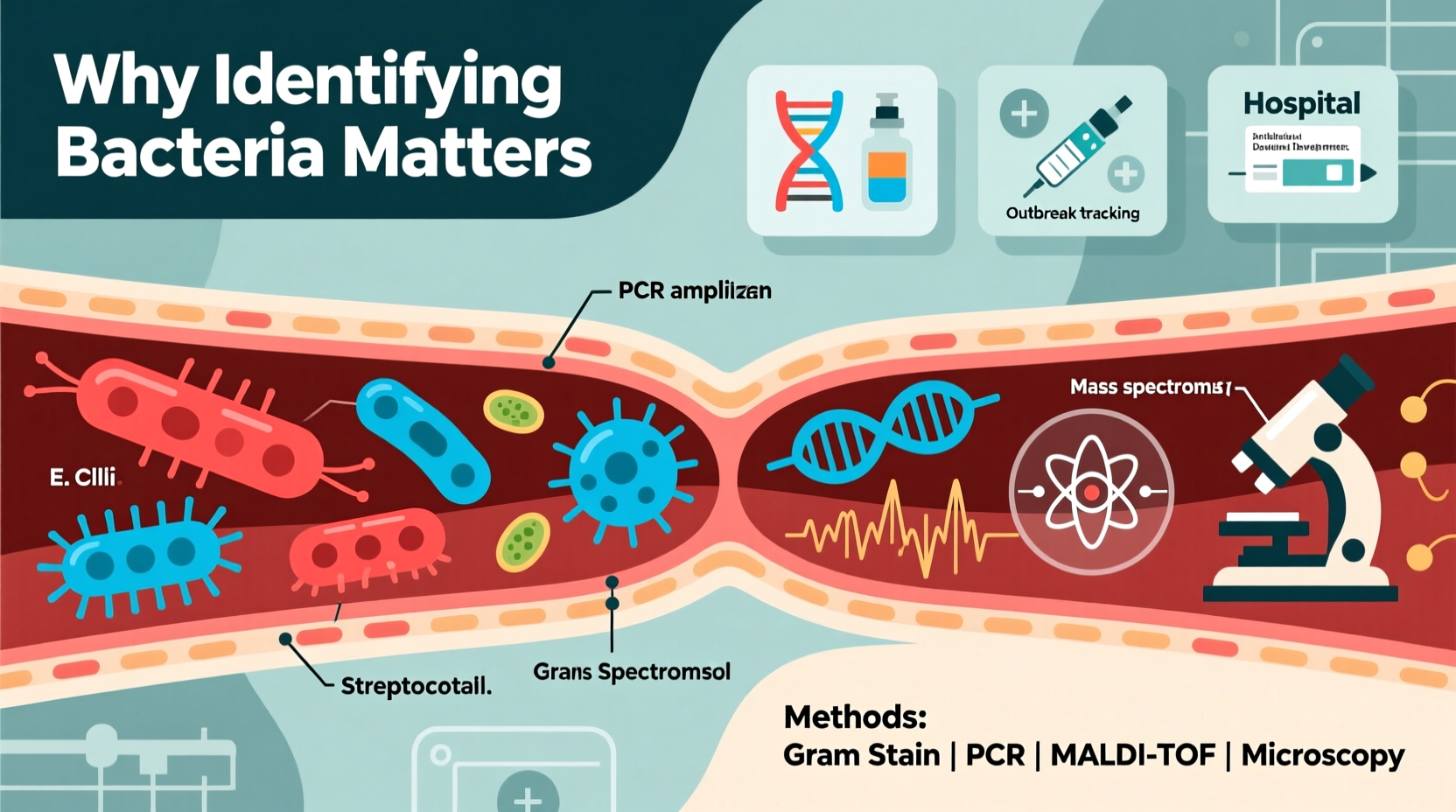
Accurate bacterial identification directly influences outcomes across multiple fields. In clinical medicine, misidentifying a pathogen can lead to inappropriate antibiotic use, treatment failure, and the rise of drug-resistant strains. For example, mistaking Staphylococcus aureus for a less virulent species could delay life-saving interventions in cases of sepsis.
Beyond human health, bacterial identification plays a vital role in food safety. Contamination by Salmonella, Listeria, or E. coli O157:H7 can trigger widespread outbreaks if not detected early. Rapid identification allows manufacturers to recall products before they reach consumers, preventing illness and protecting brand integrity.
In environmental science, knowing which bacteria are present helps assess ecosystem health. Nitrogen-fixing bacteria in soil support agriculture, while sulfate-reducing bacteria in pipelines accelerate corrosion. Monitoring microbial communities enables better management of wastewater treatment plants, oil spills, and agricultural runoff.
“Knowing the exact bacterium involved changes everything — from treatment strategy to containment protocol.” — Dr. Lena Patel, Clinical Microbiologist, Johns Hopkins Hospital
Core Methods Used in Bacterial Identification
Modern microbiology employs a tiered approach to bacterial identification, combining classical techniques with advanced molecular tools. Each method offers different levels of speed, accuracy, and cost-efficiency.
1. Morphological and Biochemical Testing
This traditional approach involves culturing bacteria on selective media and observing colony characteristics such as color, shape, and hemolysis. Gram staining distinguishes between Gram-positive and Gram-negative organisms based on cell wall structure.
Biochemical tests like catalase, coagulase, oxidase, and carbohydrate fermentation profiles help narrow down species. For instance, a catalase-positive, coagulase-positive coccus likely indicates Staphylococcus aureus.
2. Automated Culture Systems
Hospitals often use automated systems like VITEK® or BD Phoenix™, which analyze metabolic activity across dozens of substrates simultaneously. These platforms provide species-level identification within 4–24 hours after isolation.
3. Molecular Techniques
Polymerase Chain Reaction (PCR) amplifies specific DNA sequences unique to certain bacteria. Real-time PCR allows quantification and detection in under two hours. Multiplex panels can screen for dozens of pathogens at once — crucial during outbreaks.
16S rRNA gene sequencing is considered the gold standard for identifying unknown isolates. This highly conserved gene contains variable regions that serve as molecular barcodes for bacterial classification.
4. Mass Spectrometry (MALDI-TOF)
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) MS analyzes protein fingerprints from whole bacterial cells. It can identify most clinically relevant species in minutes, revolutionizing diagnostic labs since its adoption in the 2010s.
5. Whole Genome Sequencing (WGS)
WGS provides the most comprehensive data, enabling precise strain typing, virulence assessment, and antimicrobial resistance prediction. Though still costly for routine use, it's indispensable in outbreak investigations and research.
Comparative Overview of Identification Methods
| Method | Time Required | Accuracy | Cost | Best Use Case |
|---|---|---|---|---|
| Gram Stain + Culture | 24–72 hours | Moderate | Low | Preliminary diagnosis in resource-limited settings |
| Automated Biochemical Systems | 6–24 hours | High | Medium | Hospital labs for routine pathogen ID |
| MALDI-TOF MS | Minutes–hours | Very High | High (initial), Low (per test) | High-volume clinical laboratories |
| PCR / Multiplex Panels | 1–4 hours | High | Medium–High | Rapid detection without culture |
| Whole Genome Sequencing | 1–3 days | Extremely High | Very High | Outbreak tracing, research, AMR profiling |
Real-World Application: A Hospital Outbreak Investigation
In 2022, a pediatric intensive care unit in Ohio experienced a sudden cluster of bloodstream infections. Initial cultures suggested Klebsiella pneumoniae, but antibiotic resistance patterns varied unexpectedly. Using MALDI-TOF, all isolates were confirmed as the same species. However, suspicion remained due to differing clinical courses.
The hospital collaborated with the CDC to perform whole genome sequencing. Analysis revealed two distinct strains of K. pneumoniae — one carrying the NDM carbapenemase gene, making it nearly untreatable. This discovery triggered immediate infection control measures: cohorting patients, enhanced disinfection, and staff screening.
Within three weeks, new cases dropped to zero. Without precise identification at the genetic level, the resistant strain might have spread unchecked, leading to higher mortality and prolonged facility closure.
Action Plan: Steps to Improve Bacterial Identification Practices
Whether in a clinic, lab, or field setting, following a structured workflow enhances reliability and response time.
- Collect specimens properly: Use sterile techniques and appropriate transport media to avoid contamination or degradation.
- Perform initial microscopy: Gram stain provides immediate clues about bacterial morphology and guides preliminary therapy.
- Culture on selective media: Isolate colonies for further testing; use chromogenic agars for faster presumptive ID.
- Apply rapid diagnostics: Use MALDI-TOF or PCR if available to reduce turnaround time.
- Confirm with molecular methods: When dealing with outbreaks or multidrug-resistant organisms, employ sequencing for definitive analysis.
- Integrate data with electronic health records: Link results to patient histories for better surveillance and stewardship.
Frequently Asked Questions
Why can’t we rely only on symptoms to diagnose bacterial infections?
Symptoms like fever, inflammation, or fatigue are non-specific and overlap across viral, fungal, and bacterial conditions. Only direct identification of the causative agent ensures correct treatment. Treating presumed bacterial infections without confirmation contributes to antibiotic misuse.
Can bacteria be identified without culturing them?
Yes. Molecular methods like PCR and metagenomic sequencing can detect bacterial DNA directly from samples such as blood, urine, or environmental swabs. These culture-independent approaches are especially useful when organisms are slow-growing, fastidious, or killed by prior antibiotic exposure.
How does bacterial identification impact antibiotic resistance?
Precise identification allows targeted therapy instead of broad-spectrum antibiotics. This reduces selection pressure that drives resistance. Additionally, identifying resistance genes through genomic methods supports antimicrobial stewardship programs and informs public health alerts.
Conclusion: Turning Knowledge Into Protection
Identifying bacteria is far more than an academic exercise — it's a frontline defense in medicine, food safety, and environmental protection. As global challenges like antimicrobial resistance and emerging pathogens grow, the need for accurate, timely, and accessible identification methods becomes ever more urgent.
Investing in robust diagnostic infrastructure, training skilled personnel, and adopting innovative technologies aren't optional upgrades — they're essential components of a resilient public health system. Whether you're a clinician choosing the right antibiotic, a regulator ensuring safe water, or a researcher tracking microbial evolution, your decisions depend on knowing exactly what bacteria you're facing.









 浙公网安备
33010002000092号
浙公网安备
33010002000092号 浙B2-20120091-4
浙B2-20120091-4
Comments
No comments yet. Why don't you start the discussion?